By Khushi Vora, Mehul Gupta, and Sunand Kannappan
Introduction
Acute myeloid leukemia is a type of cancer caused by the formation of abnormal, undifferentiated, myeloblasts (a type of white blood cell), red blood cells or platelets. In leukemia, these immature blood cells, commonly referred to as “blast cells”, are overproduced and disseminated into the bloodstream. These blast cells do not mature into normal bloods cells but instead develop abnormally, crowding out normal blood cells and prevent them from functioning.
The signal transducer and activator of transcription 3 (STAT3) protein is a key constituent of the STAT family of proteins that has been shown to cause oncogenesis via overexpression in a variety of cancer types through different pathways. Previous work has shown that STAT 3 may be a key transcriptional regulator of TP53, which has been linked to the development of various cancer types. Additionally, preliminary work has also been done to show that STAT 3 expression is vital to leukemic blast survival, and therefore the pathology of this acute myeloid leukemia. This protein is active in the nucleus, through phosphorylation or other related chemical interactions, binding to specific areas of DNA. STAT3 acts as a transcription factor which regulates a variety of genes vital in cell growth and apoptosis, and therefore represents an important driver of cellular processes. Therefore, by identifying whether STAT3 is overexpressed in acute myeloid leukemia may offer novel insight into the parthenogenesis of this cancer. Further, developing a unique small molecule drug for STAT3, may allow for targeted inhibition of this key protein in types of cancer where STAT3 is suspected to be a driver.
OBJECTIVE
There are two major objectives for this experiment:
1. Determine if the STAT3 protein is overexpressed in acute myeloid leukemia.
2. Create a drug that can successfully bind to the STAT3 protein in only tumor cells and inhibit its function.
HYPOTHESIS
We hypothesize that the STAT3 protein will be overexpressed in acute myeloid leukemia due to the fact that it is transcription factor for multiple other oncogenic genes as well as being linked to other types of cancer. Additionally, we hypothesize that the drug being developed will inhibit the STAT3 protein. If successful, future work should attempt to look at if this drug is able to down regulate STAT3 protein expression, and therefore reduce the growth of cancer cells by slowing down transcription of genes causing the proliferation of abnormal blood cells.
MATERIALS
• PubMed • cBioPortal
• UniProt • SWISS-MODEL
• MetaPocket 2.0 • PyMOL
• E-LEA3D • SwissADME
METHODS
1. Firstly, we did a literature search, looking through PubMed articles to find more information on acute myeloid leukemia and identify potential protein targets vital to cancer pathology.
2. After substantial review of the literature, the STAT 3 protein was selected due to its activity in the TP53 pathway (Niu G, Mol Cel Biol 2005) as well as the role of the STAT 3 protein to leukemic blast survival (Redell, Blood 2011).
3. Using data from the TCGA provisional acute myeloid leukemia database on cBioPortal the mRNA expression levels of the STAT3 protein for 173 patients was obtained.
4. Using the TCGA provisional acute myeloid leukemia database on cBioPortal, the overall survival of the patients in months was also obtained.
5. After obtaining the data, we then organized it into the proper columns so that each patient had both an expression value for STAT3 and an overall survival statistic. We remove any patients that had no expression values or no overall survival data, leaving 109 patients in the study.
6. We then divided the patients into two halves based on their mRNA expression of STAT3: high expression - defined as Reads Per Kilobase of transcript per Million mapped reads (RPKM) > 0.5654 – and low expression defined as RPKM </= 0.5654. There were 55 patients in the high expression category and 54 in the low expression category
7. After this, we moved on to developing a Kaplan Meier curve which graphs the overall survival of patients in each of these groups and reports the significance value (p-value) for the difference between these two. A p-value less than 0.05 is used as the barrier of significance in this study.
8. After identifying the significance of the protein in acute myeloid leukemia, we started the drug developing process by using UniProt to find the standard amino acid sequence of the STAT3 protein which was needed to make the model of the protein.
9. The sequence was placed into Swiss Model which created the actual structure using a process called homology based protein modelling which compares the amino acid sequence queried with known 3D structures to approximate the structure of the protein.
10. Metapocket 2.0 was then used to find the five largest binding sites which would be the most effective to develop a drug for, based on their free energy of binding (a prediction as to how spontaneous the reaction will be.
11. After ranking these biding sites, we selected the binding site offering the largest free energy of binding as this is hypothesized to offer the most spontaneous reaction with the small molecule drug that we will be developing.
12. Using e-Lea3D we placed specific criteria in so that the drug then created would fit the binding site. The molecular weight was limited to less that 350 grams/mol as this is usually thought to be optimal for absorption into the body. The number of rotatable bonds was between the ranges of 2-7 because we did not want to create a drug with many conformations which would be more likely to have nonspecific binding to our target and be more computationally taxing. The XLogp which is the compounds solubility in water and lipid is 3.5 because the drug needs to be able to be absorbed into the bone marrow and blood.
13. After this, we measured the toxicity levels of the drug in silico using SwissADME to make sure that, at least in theory, this drug would be relatively non-toxic for individual consumption.
14. Last step for the drug development involved the docking analysis of the drug to see if it binds well to the STAT3 protein. This process was also conducted using e-Lea3D.
15. Finally, the small molecule and the protein were visualized with PyMol.
OBSERVATIONS
The Kaplan Meier curve created from clinical data provided the p-value of 0.774, which did not meet the barrier of significance in this case (Figure 1).
Figure 1. Kaplan Meier curve showing the overall survival of the high expression group (blue) and low expression group (green) of STAT3. The cut-off for expression in this case was 0.5654, with 55 patients in the high and 54 patients in the low expression category. The p-value associated with this graph is 0.774, indicating no statistically significant difference between the high and low treatment groups.
However, due to the importance of STAT3 in a variety of other oncogenic pathways and cancer types, we still embarked on the development of a small molecule drug that would be able to inhibit this protein. Using the methodology discussed previously, we developed a small molecule inhibitor (Figure 2) which contains elements carbon, helium, nitrogen and sulfur.
Figure 2. Visualization of STAT 3 inhibitor 1.
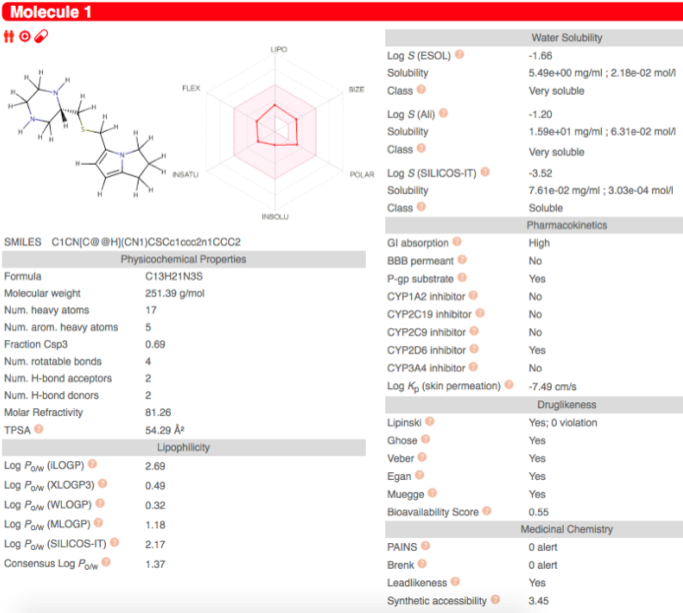
The toxicity result of the drug from SWISS-ADME shows it to be very soluble in the body, it also has a high Gastrointestinal Track (GI) absorption and the drug was not blood brain barrier permanent. These are important as it would allow the drug to have high uptake. Under the drug-likeness criteria there are no violations for general rules of effective pharmaceuticals like Lipinski, Ghose, Veber, Egan and Muegge. The synthetic accessibility of the drug was 3.45, which means it is inexpensive and easy to produce. Relative score offered by SWISS ADME indicating that the drug is relatively simple and synthesizable via known reaction mechanisms from readily purchasable compounds. These results give us relative confidence that the drug will be synthesizable in silico. Lastly the docking process showed that the drug was readily able to bind to the protein in the correct area, furthering our confidence that the drug may be a good candidate for further study.
Figure 3. Swiss ADME output for the small molecule drug created. Showcases a variety of mathematically derived parameters associated with the drug including solubility, synthetic accessibility and leadlikness.
DISCUSSION
STAT3 protein is transcription factor that is overexpressed in a variety of cancer types, and so was a candidate well suited to study for its effects on acute myeloid leukemia. Using clinical data from the TCGA Provisional database in cBioPortal, clinical data was accessed for 109 patients and a Kaplan Meier curve was developed to determine the effect of STAT3 expression and overall survival. The p-value from the Kaplan Meier curve was 0.774 which is not statistically significant. However, due to the low sample size as well as limitations of the dataset, further investigation of the role of STAT3 is worth considering. There may be mechanisms other than overexpression, such as increased phosphorylation or chemical activation via cytokines (Nguyen P, J Interferon Cytokine Res. 2015), that cause it to contribute to this disease. Though this protein appears to not be significantly associated with patient survival in acute myeloid leukemia, we determined that it would still be worthy target due to its oncogenic properties in a variety of other cancers as well as metastasis. The drug we have created is able to be easily absorbed into the body which is necessary to access and target cancerous cells overproducing this protein. Results have also shown it to be easily synthesisable making not too expensive for production. Toxicity result of the drug were ideal results; it has high solubility meaning that it would not be difficult to reach the STAT3 protein through lipids and water which is necessary to reach the cancer site. The also had a high GI absorption which indicated that it could be simply absorbed through some sort of oral medication, so it can transfer to all areas of body and be accessible to cancerous regions. The drug is also not blood brain barrier permanent which is preferential in this case, as acute myeloid leukemia and many of the other disease STAT3 are associated with exist primarily in the bone marrow. The synthetic accessibility is 3.45 so it can easily be synthesised and would not be too expensive to make which is a positive outcome. Docking result showed how effectively the drug was able to bind to the binding site and the drug we created is 37.6%. Although it does not lie in the high range of extremely successful outcome, the percentage indicates that there binds well to the protein which is the result we are looking for. A key limitation of such computational drug studies is the specificity of molecular binding, as it is not currently feasible to do large scale in silico tests to validate how readily the molecule binds to other protein targets. Therefore, in vitro and in vivo murine models are suggested for further validation of drug efficacy.
CONCLUSIONS
Though the STAT3 protein was found to not significantly regulate patient survival, it still constitutes a valid target for the development of a novel pharmaceutical. The drug that we have created has low toxicity having minimal impacts on patient’s quality of life and it can be easily synthesised meaning it will not be too costly to produce. The drug has shown to be easily absorbed by the body and violated no criteria for effective pharmaceuticals according to conventionally accepted theoretical rules. The hypothesis that we can computationally develop a drug targeting STAT 3 can therefore be accepted. The drug binds well to the overexpressed STAT3 protein. This may potentially help control the proliferation of abnormal blood cells by properly regulating the transcription of a variety of other oncogenes. Overall the computational results for the drug show it to have a potential of being effective.
REFERENCES
Acute myelogenous leukemia statistics - Canadian Cancer Society. (n.d.). Retrieved from http://www.cancer.ca/en/cancer-information/cancer-type/leukemia-acute-myelogenous-aml/statistics/?region=on
Adult Acute Myeloid Leukemia Treatment. (n.d.). Retrieved from https://www.cancer.gov/types/leukemia/patient/adult-aml-treatment-pdq
European Bioinformatics InstituteProtein Information ResourceSIB Swiss Institute of Bioinformatics. (n.d.). European Bioinformatics Institute. Retrieved from http://www.uniprot.org/
Figure 2f from: Irimia R, Gottschling M (2016) Taxonomic revision of Rochefortia Sw. (Ehretiaceae, Boraginales). Biodiversity Data Journal 4: E7720. https://doi.org/10.3897/BDJ.4.e7720. (n.d.). doi:10.3897/bdj.4.e7720.figure2f
Mencalha, A. L., Corrêa, S., Salles, D., Du, B., Santiago, M. F., & Abdelhay, E. (2014, November 23). Inhibition of STAT3-interacting protein 1 (STATIP1) promotes STAT3 transcriptional up-regulation and imatinib mesylate resistance in the chronic myeloid leukemia. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/25417721
(n.d.). Retrieved from http://www.cbioportal.org/
(n.d.). Retrieved from http://projects.biotec.tu-dresden.de/metapocket/
(n.d.). doi:10.3897/bdj.4.e7720.figure2fLahortiga, I., & Cools, J. (2012, June). Retrieved May 26, 2018, from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3366641/
Nguyen P, Putoczki T, Ernst M. STAT3-Activating Cytokines: A Therapeutic Opportunity for Inflammatory Bowel Disease?. Journal of Interferon & Cytokine Research. 2015;35(5):340-350.
Niu G, Wright K, Ma Y, Wright G, Huang M, Irby R et al. Role of Stat3 in Regulating p53 Expression and Function. Molecular and Cellular Biology. 2005;25(17):7432-7440.
Redell M, Ruiz M, Alonzo T, Gerbing R, Tweardy D. Stat3 signaling in acute myeloid leukemia: ligand-dependent and -independent activation and induction of apoptosis by a novel small-molecule Stat3 inhibitor. Blood. 2011;117(21):5701-5709.
Stone, R. M. (n.d.). The difficult problem of acute myeloid leukemia in the older adult. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/12469764
SWISS-MODEL | Home Page. (n.d.). Retrieved from https://swissmodel.expasy.org/
Treatments for acute myelogenous leukemia - Canadian Cancer Society. (n.d.). Retrieved from http://www.cancer.ca/en/cancer-information/cancer-type/leukemia-acute-myelogenous-aml/treatment/?region=on
What is acute myelogenous leukemia? - Canadian Cancer Society. (n.d.). Retrieved from http://www.cancer.ca/en/cancer-information/cancer-type/leukemia-acute-myelogenous-aml/acute-myelogenous-leukemia/?region=on
About the Author
My name is Khushi Vora and I am a young girl passionate about sciences related to medicine and human physiology. I enjoy activities such painting and spending time with friends and family as well as participating in Model UN. I find research pursuing medical cures fascinating and hope to work towards the path of helping others through a medical career. During my research, the challenges that I’ve encountered have been truly unforgettable making the results that much more rewarding. This drug development project has been a truly monumental experience in my life so far because of the amount of knowledge I was able to gain and people I have been lucky enough to have met. Although I am quite young, I believe that with inspiration, hard work, and mentorship I hope to be able to work towards more new discoveries in medicine, in the near future.